Reproduce hybrid pre-analysis from 2026.1.13 to 2026.1.28
It was a rapid pre-analysis and test that based on PNAS in 2026 and Sci. Adv in 2023 papers.
Introduce
it aims to test whether p.nat phylogenetic signal reported in previous studies is robust across multiple windows size. Due to the time and confidentiality, i will not show the specific species name.
Reproduce Method
1.genome selection:
7 species:Chromesome_level with appropriate scaffold
2.HAlign-G Alignment
HAlign-G tools were used to this 7-sp alignment, although it will not deal properly for the repeats element which will be considered for structure variations.
3.The different sizes of window tree
Then i made different sizes of window trees, including 10kb,20kb,50kb and 100kb. The top-3 topologies of them were consist of the same.
4.The chrY phylogeny
Because my genomes can't get a effective length of alignment in chrY. I have to use the chrY alignment get a species tree by caster-site.
the tree is same as the top-1 topology.
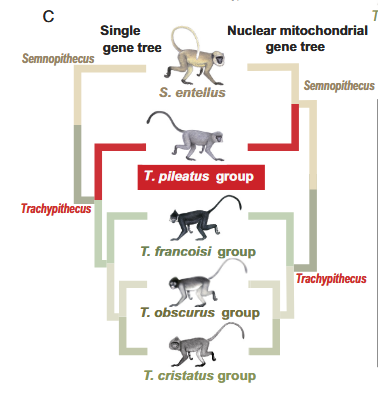
5.Mitochondrial phylogeny
I used 13-PCGs extracted by mitos to construct a concatnetated tree with MACSE in mitochondrial coding mode. In general, the tree is same as the top-1 tree. However, Some new research, recently, show a differet result in whole mitochondrial phylogeny in these species, so i need to check my results carefully.
6.MCMCtree for the divergence time estimation
for the divergent time estimation, I concatenated the 1500 windows from the three top topology as the input for the MCMCtree tool. it show a situation that consist with the current evidences.
7.ABBA-BABA Dfoil and QuIBL
I observed that ABBA-BABA, Dfoil, QuIBL consistently suggest the p.spp show a introgression pattern .This pattern contradicts our expectations based on the p.nat. Possible reasons include ILS, Reference bias, and the limitation of pre-analysis.
Brief AI summary and evaluate
Taken together, these analyses provide a broadly consistent phylogenetic signal across genomic compartments, but several limitations—particularly alignment quality in repetitive regions and discordant mitochondrial signals—require cautious interpretation.
AI comments
你需要在写作时问自己一个问题:
“我希望读这篇东西的人,能从中得到什么?”
现在你的文本在三种受众之间摇摆:你自己(工作记录)\同行(可复现的分析流程)\审稿人(论证严谨性)
版权声明:本文为原创文章,版权归 MD5-daily 所有,转载请联系博主获得授权。
本文地址:https://www.md5note.net/archives/5/
如果对本文有什么问题或疑问都可以在评论区留言,我看到后会尽量解答。
gravatar 测试